
The ferrn package extracts key components from the data object collected during projection pursuit (PP) guided tour optimisation, produces diagnostic plots, and calculates PP index scores.
You can install the development version of ferrn from GitHub with:
# install.packages("remotes")
remotes::install_github("huizezhang-sherry/ferrn")The data object collected during a PP optimisation can be obtained by
assigning the tourr::annimate_xx() function a name. In the
following example, the projection pursuit is finding the best projection
basis that can detect multi-modality for the boa5 dataset
using the holes() index function and the optimiser
search_better:
set.seed(123456)
holes_1d_better <- animate_dist(
ferrn::boa5,
tour_path = guided_tour(holes(), d = 1, search_f = search_better),
rescale = FALSE)
holes_1d_betterThe data structure includes the basis sampled by the
optimiser, their corresponding index values (index_val), an
information tag explaining the optimisation states, and the
optimisation method used (search_better). The
variables tries and loop describe the number
of iterations and samples in the optimisation process, respectively. The
variable id serves as the global identifier.
The best projection basis can be extracted via
library(ferrn)
library(dplyr)
holes_1d_better %>% get_best()
#> # A tibble: 1 × 8
#> basis index_val info method alpha tries loop id
#> <list> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <int>
#> 1 <dbl [5 × 1]> 0.914 interpolation search_better NA 5 6 55
holes_1d_better %>% get_best() %>% pull(basis) %>% .[[1]]
#> [,1]
#> [1,] 0.005468276
#> [2,] 0.990167039
#> [3,] -0.054198426
#> [4,] 0.088415793
#> [5,] 0.093725721
holes_1d_better %>% get_best() %>% pull(index_val)
#> [1] 0.9136095The trace plot can be used to view the optimisation progression:
holes_1d_better %>%
explore_trace_interp() +
scale_color_continuous_botanical()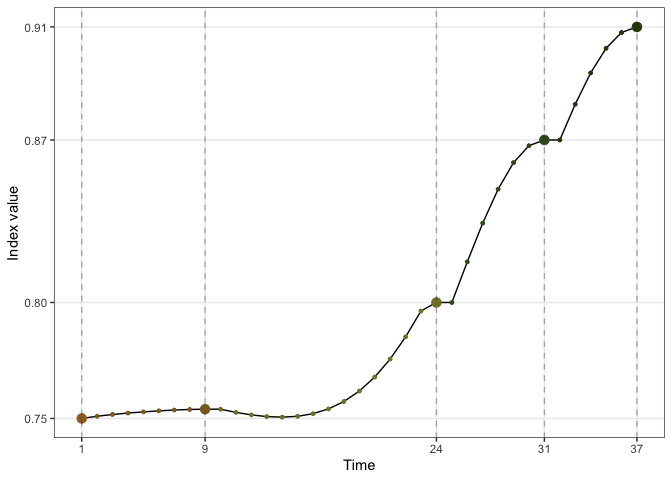
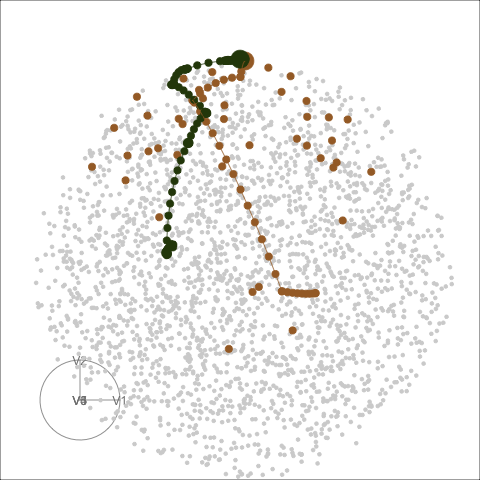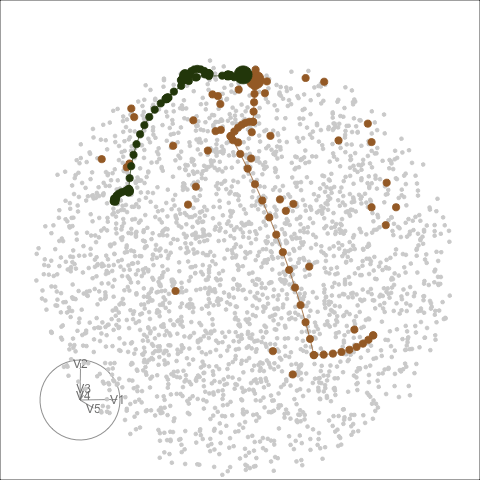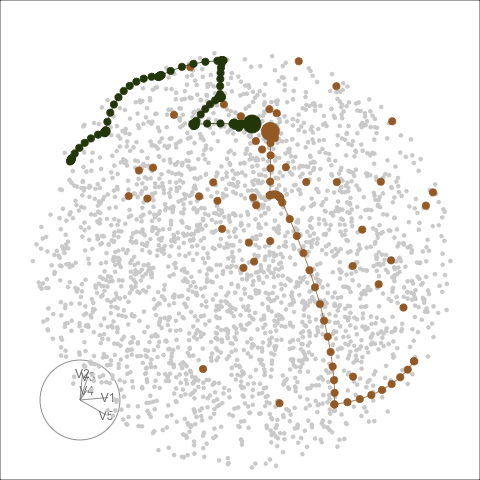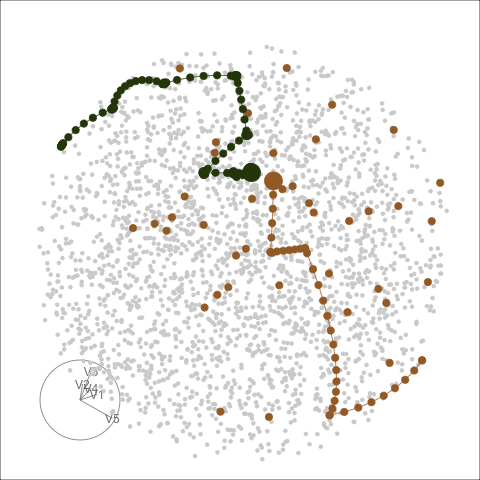
Different optimisers can be compared by plotting their projection
bases on the reduced PCA space. Here holes_1d_geo is the
data obtained from the same PP problem as holes_1d_better
introduced above, but with a search_geodesic optimiser. The
5 \(\times\) 1 bases from the two
datasets are first reduced to 2D via PCA, and then plotted to the PCA
space. (PP bases are ortho-normal and the space for \(n \times 1\) bases is an \(n\)-d sphere, hence a circle when projected
into 2D.)
bind_rows(holes_1d_geo, holes_1d_better) %>%
bind_theoretical(matrix(c(0, 1, 0, 0, 0), nrow = 5),
index = tourr::holes(), raw_data = boa5) %>%
explore_space_pca(group = method, details = TRUE) +
scale_color_discrete_botanical()
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the ferrn package.
#> Please report the issue at
#> <https://github.com/huizezhang-sherry/ferrn/issues>.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.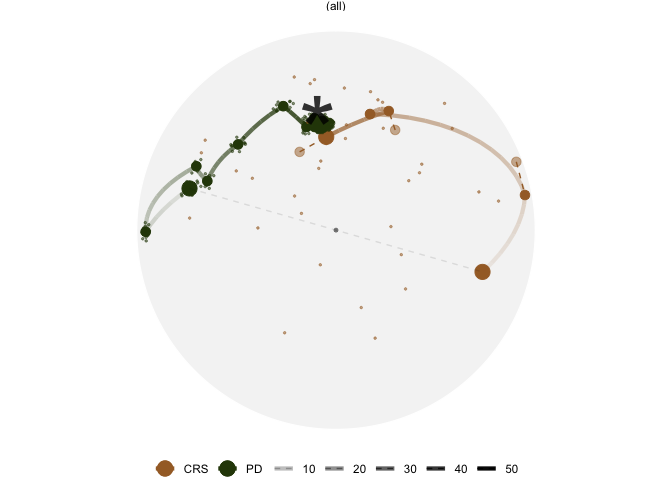
The same set of bases can be visualised in the original 5-D space via tour animation:
bind_rows(holes_1d_geo, holes_1d_better) %>%
explore_space_tour(flip = TRUE, group = method,
palette = botanical_palettes$fern[c(1, 6)],
max_frames = 20,
point_size = 2, end_size = 5)